Zebrafish and humans have new biomedical friend in the spotted gar
 The Spotted Gar.: image credit: Solomon DavidThe genome of a slowly evolving fish, the spotted gar, is so much like both zebrafish and humans that it can be used as a bridge species that could open a pathway to important advancements in biomedical research focused on human diseases.
The Spotted Gar.: image credit: Solomon DavidThe genome of a slowly evolving fish, the spotted gar, is so much like both zebrafish and humans that it can be used as a bridge species that could open a pathway to important advancements in biomedical research focused on human diseases.
That is the conclusion of a comprehensive and international research effort, led by the University of Oregon in collaboration with a number of international partners including Bangor University. Researchers sequenced the genome of the gar (Lepisosteus oculatus) – an ancient fish with hard diamond-shaped scales and a long mouth filled with needle-like teeth. Their work is detailed in a paper published online in the journal Nature Genetics.
As the data were analyzed, researchers realized the spotted gar’s genome is an evolutionary repository of ancient genetic materials, said UO biologist John H. Postlethwait. “Many genes found in human but not in zebrafish, a favorite for biomedical research, are present in gar, and, likewise, genes present in zebrafish but not in humans are also in gar,” he said. “This relic of a fish retains ancestral characteristics lost by other fishes or humans.”
Dr John Mulley, from the School of Biological Sciences at Bangor University uncovered one particularly interesting example of this process, where gar had retained an additional copy of a gene involved in pancreas development and insulin regulation (Pdx2).
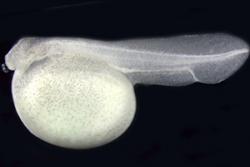 Four- day old spotted gar embryo.: Image credit: Ingo Braasch.“It’s particularly exciting for me to see this genome sequence published, as my involvement in the gar work goes back almost ten years, to my PhD research, where I travelled to Louisiana several times to participate in the captive breeding projects and carried out a range of early genome comparisons between gar and other ray-finned fish.”
Four- day old spotted gar embryo.: Image credit: Ingo Braasch.“It’s particularly exciting for me to see this genome sequence published, as my involvement in the gar work goes back almost ten years, to my PhD research, where I travelled to Louisiana several times to participate in the captive breeding projects and carried out a range of early genome comparisons between gar and other ray-finned fish.”
Numerous evolutionary conserved, non-coding genetic elements are found in gar that are often tied to human diseases, but are undetectable in zebrafish directly. “The bottom line is that the sequences of non-coding elements have been conserved in humans, gar and zebrafish -- even though they don’t code for a protein, they have to be doing something important,” said Postlethwait. “Now, if you first compare human to gar, and then gar to zebrafish, then we can make the connections. The gar is a bridge.”
The genetic importance is traced to some 450 million years ago, when bony vertebrates split into two major groups. Lobe-finned fishes went one way and ray-finned fishes another. Humans and other limbed creatures emerged along the lobe-finned line. The ray-finned line resulted in so-called teleost fishes that are the most common fish recognizable today: zebrafish, stickleback, salmon, tuna, halibut and about every fish found in a home aquarium.
Gars split off the ray-finned evolutionary line before a genome duplication gave rise to the teleost fishes. The researchers concluded that the spotted gar, by missing that duplication, maintained a conserved genetic makeup, including many entire chromosomes similar to those in the ancestor of bony vertebrates.
“By illuminating the legacy of genome duplication, the gar genome bridges teleost biology to human health, disease, development, physiology and evolution,” the researchers wrote in the paper.
The researchers report that gars are well-suited as genomic and laboratory models. Other non-teleost ray-finned fish groups – bichirs, sturgeons, paddlefish, and bowfin – are problematic due to genetic alterations that have occurred during the evolution of their individual lineages or problems with growing them in the lab. In comparison, it is relatively easy to rear gars in a lab and get embryos for developmental studies.
Gars today are found in U.S. states along the Gulf of Mexico, up the Mississippi River to Michigan in North America, as well as in Central America and Cuba. Spotted gars collected for the research were captured in Louisiana in collaboration with Allyse Ferrara of Nicholls State University.
The spotted gar genome offers a window into the evolution of vertebrate body plans, including for example how fins evolved into limbs that allowed fish to walk onto land. In addition, gars have enamel-bearing teeth and strong scales containing ganoin, which has long been thought to be a type of enamel similar to that found on human teeth. The group’s gene activity studies suggest that our teeth originated from genetic programs that formed protective scales in the skin of ancient armored fish.
The Nature Genetics paper was co-authored by 61 researchers from 33 institutions.
The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons, DOI: 10.1038/ng.3526
Publication date: 9 March 2016
